Partners: INSTRUCT-ERIC, EU-OpenScreen/Fraunhofer ITMP and EuroBioImaging
Project description
This project represents the effort of 3 EOSC-Life partners to propel the imaging data within the European scientific landscape towards embracing the FAIR principles—making data Findable, Accessible, Interoperable, and Reusable. The project focused on sharing and integrating imaging data in various fields, with a special spotlight on fighting SARS-CoV-2.
By developing and extending four distinct, open-access infrastructures, the initiative aimed at connecting different domains of the imaging community, from cellular to molecular levels.
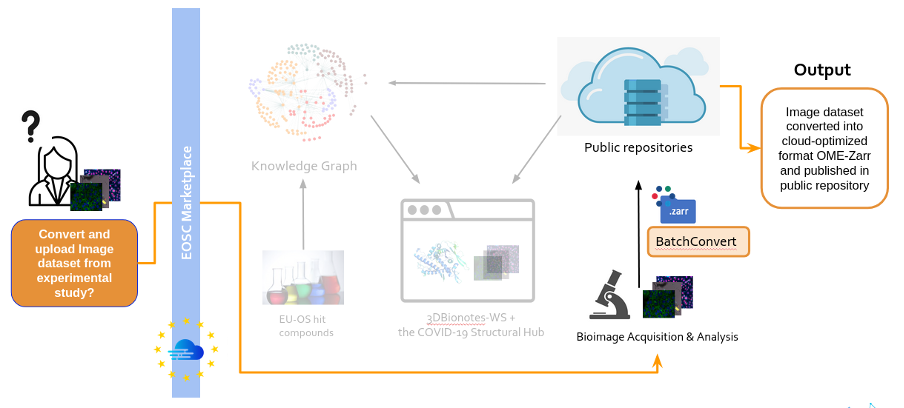
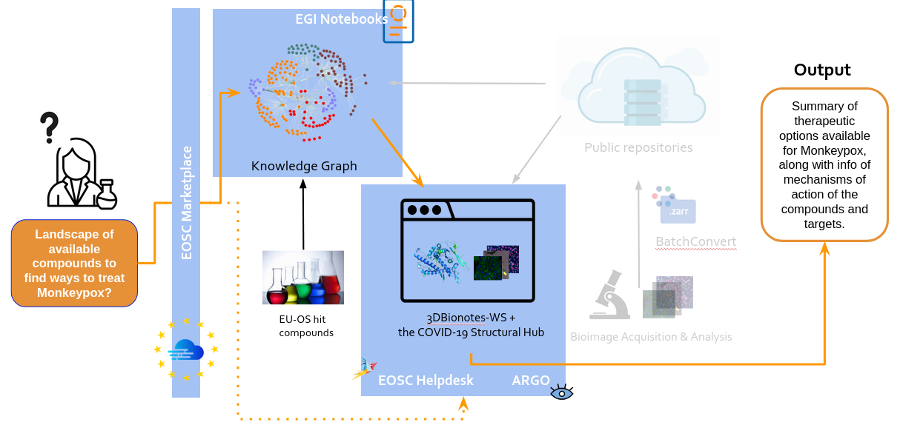
BatchConvert enables the concurrent conversion of image data collections to standard cloud-optimised file formats and their transfer to/from remote storage, hence addressing the critical need for efficient large data handling. Meanwhile, COVID-19 and infectious diseases knowledge graphs by EU-OpenScreen encapsulate the molecular landscape of these diseases, aiding in the discovery of potential treatments. 3DBionotes-WS and the COVID-19 Structural Hub enhance access to protein models and their annotations, including those coming from the bioimaging field bridging the gap between bioimaging data and molecular insights.
This strategic alignment of resources and efforts was designed to provide researchers with a panoramic view of the disease, unraveling its molecular underpinnings and mapping out the therapeutic landscape. The success of this approach in the context of SARS-CoV-2 sets a scalable and adaptable blueprint for tackling other infectious diseases.
Technical challenge
The initiative addresses the critical social challenge of accelerating scientific understanding and treatment discovery for COVID-19, aiming to equip researchers with the data and tools necessary for swift, informed responses to this and future health crises.
Achieving efficient image data sharing, data harmonization and interoperability presented substantial technical hurdles, including the difficulty of handling large image datasets, lack of standardization for data and metadata and a lack of connection between high-level imaging data and molecular imaging data.
EOSC added value
EOSC Future played a pivotal role in the seamless integration of the three key infrastructures within the project, significantly streamlining communication and coordination efforts. By incorporating the project’s outputs into its centralized, user-friendly Marketplace, EOSC widened their visibility and accessibility to the research community. Insight into the impact and effectiveness of these outputs was facilitated through detailed reports from EOSC Core Services, offering valuable feedback on their contribution to resolving community challenges. Additionally, EOSC Future offered robust infrastructure support for deploying the project’s tools, ensuring efficient implementation and widespread adoption.
Main results
- BatchConvert, a command line tool capable of generating and running workflows for automated and parallelized conversion of bioimage collections into standardized, cloud-optimized file formats (OME-Zarr). The tool supports high-performance data transfer from/to remote storage as part of the executed workflows, and therefore allows for a quick and efficient submission of bioimage data to public repositories.
- Additionally, a complementary Galaxy tool has been developed. With a web-based graphical user interface, this tool enables the creation of OME-Zarr data in the Galaxy cloud computing environment, facilitating the construction of cloud-based workflows consuming the OME-Zarr format.
- COVID-19 (or other infectious disease) knowledge graph, a representation of compounds and biological entities to facilitate the understanding of the molecular basis of COVID-19 and the landscape of available compounds to treat this disease, as a result of the collection of data from fragment-based drug discovery studies and bioimaging assays from public repositories.
- 3DBionotes-WS extension and the COVID-19 Structural Hub, two web open-access platforms that act as a data aggregator to enable user-friendly access to protein models and their annotations, including post-translational modifications, genomic variations associated with diseases, short linear motifs, immune epitopes sites and those coming from bioimaging domain, among others.
Demos
Other resources
Webpages
- Galaxy tool for conversion to OME-Zarr: https://imaging.usegalaxy.eu/?tool_id=toolshed.g2.bx.psu.edu%2Frepos%2Fimgteam%2Fbioformats2raw%2Fbf2raw%2F0.7.0%2Bgalaxy2&version=latest
- 3DBionotes-WS Official Webpage: https://3dbionotes.cnb.csic.es/ws
- COVID-19 Structural Hub Official Webpage URL: https://3dbionotes.cnb.csic.es/ws/covid19
Code Sources
- BatchConvert Github: https://github.com/Euro-BioImaging/BatchConvert
- 3DBionotes-WS and COVID-19 Structural Hub Github: https://github.com/3dbionotes-community/3DBIONOTES
- COVID19 KG GitHub: https://github.com/Fraunhofer-ITMP/BY-COVID-KG
- Mpox KG GitHub: https://github.com/Fraunhofer-ITMP/mpox-kg
Publications
- BatchConvert: https://preprints.arphahub.com/article/116669/
- OME-Zarr: https://www.biorxiv.org/content/10.1101/2023.02.17.528834v4
- COVID-19 KG: https://zenodo.org/records/7351221
- Mpox KG: https://doi.org/10.1093/bioadv/vbad045
